
February 2, 2011
LA JOLLA, CA—Reprogramming adult cells to recapture their youthful “can-do-it-all” attitude appears to leave an indelible mark, found researchers at the Salk Institute for Biological Studies. When the team, led by Joseph Ecker, PhD., a professor in the Genomic Analysis Laboratory, scoured the epigenomes of so-called induced pluripotent stem cells base by base, they found a consistent pattern of reprogramming errors.
What’s more, these incompletely or inadequately reprogrammed hotspots are maintained when iPS cells are differentiated into a more specialized cell type, providing what the researchers dubbed an iPS cell-specific signature. “We can tell by looking at these hotspots whether a cell is an iPS cell or an embryonic stem cell,” says Ecker. “But we don’t know yet what it means for their self-renewal or differentiation potential.”
Their findings, published in the February 3, 2011, issue of Nature, confirm that iPS cells, which by all appearances look and act like embryonic stem cells, differ in certain aspects from their embryonic cousins, emphasizing that further research will be necessary before they can rightfully take embryonic stem cells’ place.
The fact that reprogramming of somatic (body) cells does not pose the same ethical quandaries as working with stem cells isolated from embryos prompted scientists into developing iPS technology for human cells that are just as potent as human embryonic stem cells, with the hope that one day, iPS cell technology can be applied to regenerative medicine.
But before cells derived from iPS cells can be used to repair tissue damaged through disease or injury, some remaining questions have to be solved. “Embryonic stem cells are considered the gold standard for pluripotency,” says Ecker. “So we need to know whether—and if so, how—iPS cells differ from ES cells.”
The reprogramming process, which turns back the clock and endows fully differentiated cells with pluripotent potential, is not a genetic transformation but an epigenomic one. The epigenome is what differentiates a fibroblast from a hepatocyte and a stem cell from a fully differentiated cell. With a few exceptions, every cell in our body contains the same genome, but epigenomic marks—tiny tags atop DNA that can tell your genes to turn on or off, to speak up or speak softly—determine a cell’s gene expression profile and hence its fate.
While others have compared genomic bits and pieces between iPS and embryonic stem cells—and found small differences—the Salk researchers and their collaborators at the University of Wisconsin and the University of California, San Diego, set their sights higher.
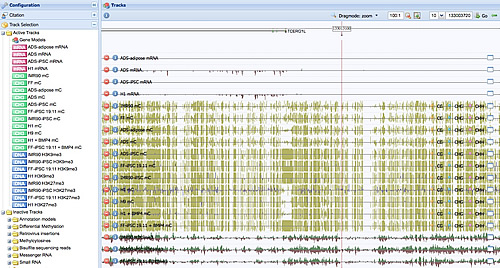 The reprogramming process leaves indelible marks in the methylation profile of induced pluripotent stem cells.
The reprogramming process leaves indelible marks in the methylation profile of induced pluripotent stem cells.
Image: Courtesy of Dr. Ryan Lister, Salk Institute for Biological Studies
They scrutinized whole-genome DNA methylation profiles—methylation is one the best-studied and most important epigenetic tags—at single-base resolution in five iPS cell lines, along with the methylomes of embryonic stem cells and somatic cells and differentiated iPS cells and differentiated embryonic stem cells.
Reprogramming induces a remarkable wholesale reconfiguration of the DNA methylation pattern throughout the genome, returning partially methylated domains to a fully methylated state; reinstating so-called non-CG methylation; and reprogramming most unmethylated and methylated CG islands, which play a crucial role in regulation gene activity, to an embryonic stem cell-like state.
“Overall, this process results in an iPS cell methylation pattern that’s very similar to that of embryonic stem cells,” says postdoctoral researcher and co-first author Ryan Lister. “But when we started to dig deeper, we discovered significant differences.”
Their experiments revealed considerable variability between iPS cell lines, including a “memory” of their tissue of origin. “Some marks carry over,” explains Ecker. “If iPS cells were derived from adipose tissue, we can see that they “remember” some methylation marks from being a fat cell.” Furthermore, new methylation patterns not found in either embryonic stem cells or the tissues of origin were identified in the iPS cells, and many of the regions showing epigenomic changes were disrupted in all iPS lines studied.
But regardless of their individual history, iPS cells showed a common defect-hotspots near telomeres and centromeres that proved resistant to reprogramming. Averaging more than one million bases in length, these hotspots failed to acquire the non-CG methylation typical of embryonic stem cells.
“These regions are really signatures,” explains postdoctoral researcher and co-first author Mattia Pelizzola. “They are shared in iPS cells derived from different parental cells, by different research groups and using different methodologies. Moreover, these regions coincide with specific modifications of histones—proteins that are important to determine the accessibility and the activity of genomic regions—and the genes contained within these regions are less expressed.”
However, when the researchers zoomed in closer, they found that the opposite held true for CG islands, short stretches of CG-rich DNA sequences that are typically found in the proximity of genes, where they may regulate gene activity. “The consequence is that some genes within these areas seem to be silenced by the altered CG island methylation patterns in the iPS cells,” says Lister. “Conceivably, these changes could limit the potential fate of the iPS cells.”
To gain a better understanding of the implications, they looked again at these regions after differentiating embryonic stem cells and iPS cells into trophoblasts, a standard cell differentiation assay. A subset of iPS cell-specific silencing marks were transmitted to differentiated cells at high frequency. “They are not easily removed,” says Lister, “and could be used as a diagnostic marker for incomplete reprogramming.”
Adds Ecker: “Now that we know that these regions exist, we want to understand why these regions can’t be reprogrammed to a more ES cell-like state.”
The research was funded in part by the California Institute for Regenerative Medicine, Catharina Foundation, American Cancer Society, Japan Society for the Promotion of Science, Mary K. Chapman Foundation, National Science Foundation, Morgridge Institute for Research and Howard Hughes Medical Institute.
Researchers who also contributed to the work include Joseph R. Nery, Ronan O’Malley and Rosa Castanon in the Genomic Analysis Laboratory at the Salk Institute; Michael Downes, Ruth Yu and Howard Hughes Medical Institute investigator Ronald M. Evans in the Gene Expression Laboratory at the Salk Institute; R. David Hawkins, Gary Hon, Sarit Klugman and Bing Ren at the Ludwig Institute for Cancer Research, University of California, San Diego; and Jessica Antosiewicz-Bourget, Ron Stewart and James A. Thomson at the Morgridge Institute for Research and the Genome Center of Wisconsin, both in Madison, Wisconsin.
About the Salk Institute for Biological Studies:
The Salk Institute for Biological Studies is one of the world’s preeminent basic research institutions, where internationally renowned faculty probe fundamental life science questions in a unique, collaborative, and creative environment. Focused both on discovery and on mentoring future generations of researchers, Salk scientists make groundbreaking contributions to our understanding of cancer, aging, Alzheimer’s, diabetes and infectious diseases by studying neuroscience, genetics, cell and plant biology, and related disciplines.
Faculty achievements have been recognized with numerous honors, including Nobel Prizes and memberships in the National Academy of Sciences. Founded in 1960 by polio vaccine pioneer Jonas Salk, M.D., the Institute is an independent nonprofit organization and architectural landmark.
Office of Communications
Tel: (858) 453-4100
press@salk.edu