
August 30, 2018
Salk scientists who discovered that aggressive breast cancer reprises pathways found in fetal cells have identified genetic master switch for process
Salk scientists who discovered that aggressive breast cancer reprises pathways found in fetal cells have identified genetic master switch for process
LA JOLLA—A team at the Salk Institute has identified a master switch that appears to control the dynamic behavior of tumor cells that makes some aggressive cancers so difficult to treat. The gene Sox10 directly controls the growth and invasion of a significant fraction of hard-to-treat triple-negative breast cancers.
Recently, the Salk lab led by Professor Geoffrey Wahl discovered that aggressive breast cancers return to a flexible, earlier state found in fetal breast tissue. This cellular reprogramming may be the key to cancer’s ability to form new cell types, evolve drug resistance and metastasize to other locations in the body. The new work documenting Sox10’s role in this process, which was reported in the journal Cancer Cell on August 30, 2018, represents a major milestone in researchers’ understanding of cancer and could open new avenues for diagnosing and treating aggressive breast cancer as well as other types of intractable cancers.
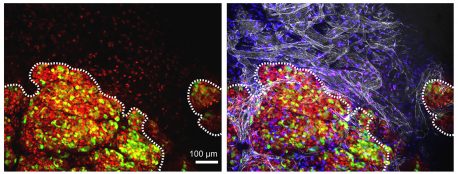
Click here for a high-resolution image.
Credit: Salk Institute/Sanford Consortium for Regenerative Medicine
“Two things that make triple-negative breast cancers so hard to treat are their heterogeneity—they have many different cell types within a single tumor—and their ability to move around and colonize new areas, the process of metastasis,” says Wahl, holder of the Daniel and Martina Lewis Chair and senior author of the work. “It’s what you could call the imprecision in precision medicine, in the sense that we might target one type of cell, but there are other cells within the tumor that can change to become drug resistant, analogous to how a chameleon changes colors to evade predators.”
In order to develop from a single cell into a complete organism such as a mouse or human, embryonic and fetal cells have the ability to divide rapidly, move throughout the body and change into multiple different cell types—properties known as “plasticity.” But adult cells turn off this plasticity, which, for reasons that aren’t fully understood, can get reawakened and turn cells cancerous.
“The embryo will supercharge certain cells to rapidly start critical developmental processes that spawn the growths of new tissues, but it’s very important that these cells get shut off when your body no longer needs this to occur,” says Christopher Dravis, a Salk staff scientist and the paper’s co-first author. “In aggressive breast cancers, we’re finding that safety mechanisms that regulate these powerful development genetic programs are lost, so these processes underlying cellular plasticity are being reactivated to drive tumor development and ultimately the malignancy that’s associated with the disease.”
In the new study, the Wahl lab began by examining which parts of mouse mammary cells’ DNA—which is tightly coiled in a package called chromatin—was uncoiling to make specific genes more accessible. This was the team’s first clue to which genes might be active during development. The chromatin analysis revealed that in both fetal cells and a subpopulation of breast tumor cells, the same areas of the genome were becoming accessible—areas where a master gene regulator called Sox10 is known to bind to DNA to initiate a variety of developmental processes. It seemed like a gene regulation “smoking gun.”

Click here for a high-resolution image.
Credit: Salk Institute
“In fetal cells, which are the most ‘plastic,’ we saw that binding sites for Sox10 were very open and accessible compared to healthy adult cells, which are mostly inflexible and the chromatin is very closed,” says Chi-Yeh Chung, a Salk research associate and the paper’s co-first author.
Next the team showed that Sox10 actually bound to genes in the open regions to activate them, thereby directly regulating genes responsible for cell type, mobility and other features relevant to breast cancer’s ability to evolve and metastasize. Breast cancer cells with high levels of Sox10 changed to become much more primitive and acquired the ability to move. The results were so dramatic that the team repeated the experiment with a technique to keep Sox10 from binding to those genes. This time, without access to Sox10, none of the breast cells that had been programmed to turn cancerous were now able to form tumors.
“When you’re talking about metastasis, a result like this is huge,” says Bianca Lundien Kennedy, a two-time breast cancer survivor and patient/research advocate who has worked with the Wahl lab for seven years. “It drastically alters the research landscape and will be such a personally significant thing for any breast cancer survivor or person with cancer to hear. It really hits home.”
The researchers caution that strategies to block Sox10 will require further development and will have to be tested for safety to determine if they impact normal cellular functions. However, the fact that Sox10 regulates many genes potentially linked to aggressive breast cancer offers the potential to target one or more of them as the basis for developing “personalized” therapies for metastatic breast cancer. Additionally, the findings could lead to diagnostic tests for breast and other cancers by checking adult tissues for proteins that normally would only be produced by fetal cells.
The team next plans to explore potential therapeutic drug targets among the genes regulated by Sox10.
Other authors included: Nikki K. Lytle and Tannishtha Reya of Sanford Consortium for Regenerative Medicine and UC San Diego Moores Cancer Center; and Jaslem Herrera-Valdez, Gidsela Luna and Christy L. Trejo of Salk.
The work was funded by the National Institutes of Health/National Cancer Institute (R35 CA197687); the Susan G. Komen Foundation (SAC110036); the Breast Cancer Research Foundation; and NIH National Research Service Award fellowships F32CA174430, GM007752, CA206416, CA186043 and CA197699.
JOURNAL
Cancer Cell
AUTHORS
Christopher Dravis, Chi-Yeh Chung, Nikki K. Lytle, Jaslem Herrera-Valdez, Gidsela Luna, Christy L. Trejo, Tannishtha Reya and Geoffrey M. Wahl
Office of Communications
Tel: (858) 453-4100
press@salk.edu
Unlocking the secrets of life itself is the driving force behind the Salk Institute. Our team of world-class, award-winning scientists pushes the boundaries of knowledge in areas such as neuroscience, cancer research, aging, immunobiology, plant biology, computational biology and more. Founded by Jonas Salk, developer of the first safe and effective polio vaccine, the Institute is an independent, nonprofit research organization and architectural landmark: small by choice, intimate by nature, and fearless in the face of any challenge.