
August 16, 2006
La Jolla, CA – In times of plenty, the uni-cellular slime mold Dictyostelium discoideum leads a solitary life munching on bacteria littering the forest floor. But these simple creatures can perform heroic developmental acts: when the bacterial food supply dries up, Dictyostelium amebas band together with their neighbors and form a multi-cellular tower designed to save the children.
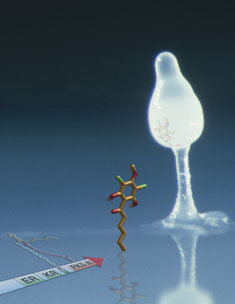
When the food supply dries up, solitary Dictyostelium discoideum cells congregate and fuse into a spore-producing tower. A newly discovered hybrid enzyme called Steely2 (shown in cartoon form) forges the basic structure of the chemical signal (DIF-1, shown here as a stick model) that orchestrates this vital step in the life cycle of Dictyostelium: the transformation of omnipotent cells into dedicated spore or stalk cells. (Image by Mike Austin using a photo by Rob Kay.)
In a forthcoming study in Nature Chemical Biology, investigators at the Salk Institute for Biological Studies and the Medical Research Council of Molecular Biology (MRC) in Cambridge, England, use traditional and computer-based methods to show how Dictyostelium synthesizes the chemical signal called DIF-1, short for Differentiation Inducing Factor, required for this developmental transformation.
The collaboration, explains co-senior author Joe Noel, Ph.D, a Howard Hughes Medical Institute investigator at Salk, “shows the power of a combined approach involving bioinformatics, enzymology, structural biology and genetics to get at the heart of why organisms exploit natural chemicals to survive and prosper in challenging ecosystems.”
When slime molds starve, they collectively form a multicellular slug-like creature that locomotes en masse to a warm spot. There, in response to the DIF-1 signal, slugs literally stand up and their cells metamorphose into either a column of stalk cells or next-generation spore cells, which perch atop the column waiting for food supplies to be restored.
Noel and Michael Austin, Ph.D., a postdoctoral fellow in Noel’s lab and co-lead author of the study, have an ongoing interest in the biosynthesis of diverse plant and microbial polyketides by enzymes known as type III PKSs. Plants make polyketide natural products such as flavonoids and stilbenes for use as sunscreens, antibiotics, flower pigments and anti-oxidants. Explains Austin, “Plant polyketides are also increasingly recognized to have significant benefits in the human diet as health-promoting components of green tea, red wine, and soybeans.”
Turns out, DIF-1 belongs to the same crowd. “While reading a review article on the diversity of naturally occurring polyketides, we realized that the core chemical structure of DIF-1, an important developmental signal in Dictyostelium, is similar to natural products made by plant type III PKSs,” recalls Austin.
At the time, Dictyostelium was in the midst of having its genome sequenced, and the bits and pieces of raw DNA sequencing data were being deposited in publicly available databases. Austin recalled, “One night I performed a bioinformatics search to look for genetic evidence that would suggest the existence of a type III PKS in Dictyostelium.” Using various computer programs to find, assemble, and translate in silico the relevant raw DNA sequencing fragments first into genes then into the proteins these genes encode, Austin reconstructed two type III PKS-like gene sequences, and also found a surprise.
Unexpectedly, these deduced genetic blueprints for type III PKSs revealed each Dictyostelium type III PKS to be fused to other enzymatically active protein domains. This never seen before hybrid arrangement works like a very efficient bucket brigade that synthesizes polyketide molecules in slime mold cells.
“Nature has paved the way to exploit this novel domain arrangement to bioengineer more efficient ways of making modified polyketides for human uses,” said Austin.
Moving to the bench, Austin and Noel lab manager Marianne Bowman isolated Dictyostelium DNA encoding the type III PKS domains and not only determined their structure, which indeed resembled a plant PKS, but also showed that one of them, called Steely2, made the chemical scaffold of DIF-1 in a test tube. All that was left was to prove was that slime molds themselves used the newly discovered enzyme to make DIF-1.
For that Noel and Austin turned to co-senior author Robert Kay, Ph.D., a Dictyostelium cellular differentiation expert and groupleader at the MRC. “We wrote a paper and sent a version to Rob Kay and said, ‘You don’t know us, but here’s what we do. Biochemically we have identified the machinery that makes the essential precursor for the bioactive DIF-1 molecule.’ “
Kay replied that he and co-lead author Tamao Saito, PhD., a scientist on sabbatical in his lab, had also focused on these unusual type III PKS genes following the recently completed final assembly and annotation of the entire Dictyostelium genome, which was carried out by a worldwide collaboration of many scientists, including the Kay group.
Working independently, Saito and Kay had deleted the Dictyostelium gene for Steely2. Not only could the resulting “deficient” slime molds not make DIF-1 but they couldn’t construct the rescue tower, which was exactly the biological corroboration that the Noel lab wanted to hear. The two labs pooled data and now publish their work as one, very complete story while continuing to collaborate on the chemical diversity found in this fascinating organism that crawls around on the forest floor.
Says Noel, who is a professor in the Jack H. Skirball Center for Chemical Biology and Proteomics at Salk, “This is a wonderful example of where egos get pushed aside about who did what and instead, as a scientific community, groups come together to address a fundamental question in biology. In the process, we collectively discovered an efficient chemical factory in Dicytostelium cells that informs us about how to modify similar systems used in other organisms to produce important medicines from nature.”
For Noel the problem is to understand biocomplexity at a level traditionally ignored – the plethora of natural chemicals found throughout nature. Organisms use chemicals as a means of interacting with their surroundings and mankind has exploited this fact to discover the vast majority of pharmaceuticals used to treat disease today. “The major fundamental question in our case is why do organisms make chemicals, what role do these molecules play in nature and how does the cellular machinery used to make them evolve over millions of years to provide new ways for the host organisms to survive and prosper. Understanding the diversity of natural chemicals and the machinery that produces them gives us a window to look back in time and understand how organisms evolve at the molecular level.”
Also contributing to the study were Steven Haydock, MD, who worked with Kay, Atsushi Kato, who along with Saito is now at Hokkaido University in Sapporo, Japan, and Bradley Moore, PhD., at Scripps Institute of Oceanography,
The Salk Institute for Biological Studies in La Jolla, California, is an independent nonprofit organization dedicated to fundamental discoveries in the life sciences, the improvement of human health and the training of future generations of researchers. Jonas Salk, M.D., whose polio vaccine all but eradicated the crippling disease poliomyelitis in 1955, opened the Institute in 1965 with a gift of land from the City of San Diego and the financial support of the March of Dimes.
Office of Communications
Tel: (858) 453-4100
press@salk.edu