Associate Professor
Laboratory of Genetics
Integrative Biology Laboratory

Human genetic differences, known as genetic variants, are what make people unique. Genetic variation affects traits like hair and eye color and can promote both health and disease. Thousands of genetic variants have now been associated with human diseases; however, the functions of most of these variants are unknown and difficult to determine, since most of them are “noncoding” and lie outside of genes (the portions of the genome that encode proteins). Research now suggests that—rather than altering genes directly—these variants instead influence gene expression by modifying when genes are turned off and on and the amount of protein that they produce in different cell types and conditions. To understand the genetic underpinnings of complex human diseases, it is essential to understand how these noncoding genetic variants alter the regulation of gene expression.
Graham McVicker’s lab studies how human genetic variation affects gene regulation by combining experimental approaches with computational analyses. The experimental side of the laboratory specializes in CRISPR perturbations of the genome, as well as in genomic assays to determine chromatin structure and gene expression. The computational side of the laboratory specializes in analysis of human genetic variation and large experimental datasets using methods from statistics and machine learning.
As a postdoctoral researcher, McVicker published the first study to identify histone mark quantitative trait loci (QTLs) in the human genome, which are genetic variants that are associated with differences in histone modifications between human individuals. He also developed a software package for analyzing allele-specific expression and discovering molecular quantitative traits, called WASP, which his lab has used to discover genes with recurrent dysregulation in cancer genomes.

McVicker’s lab has developed techniques and analysis methods that leverage CRISPR to scan noncoding portions of the genome for important regulatory functions. They also use these methods to discover important regulatory sequences that affect gene expression in T cells.
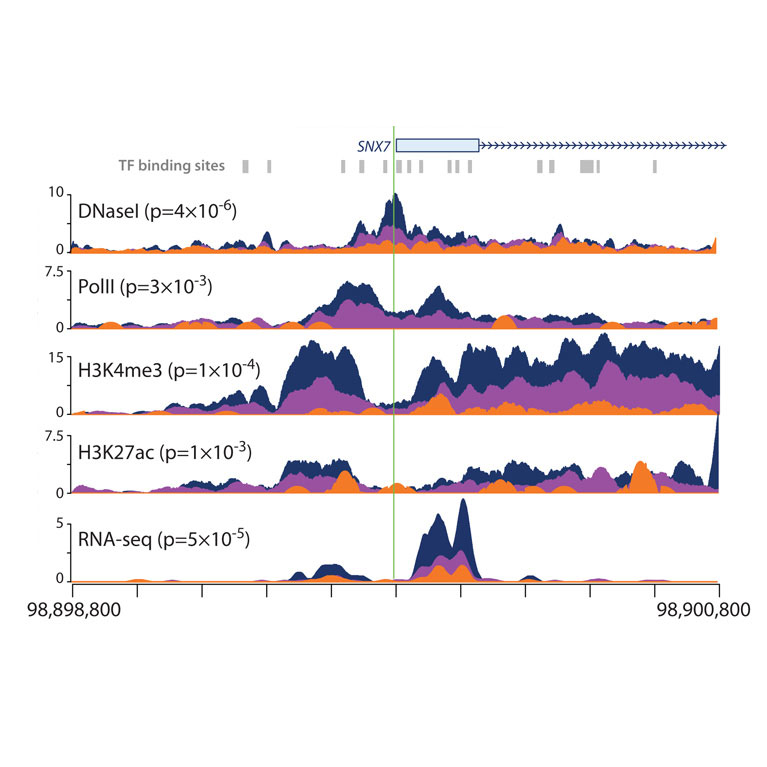
In current research, McVicker’s lab is using machine learning to predict the regulatory function of noncoding genetic variants in the human genome.
BSc Computer Science, University of British Columbia
PhD, Genome Sciences, University of Washington
Postdoctoral Scholar, University of Chicago
Postdoctoral Scholar, Stanford University